Color Atoms Pymol Zdarma
Color Atoms Pymol Zdarma. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Labeling is important so there are many options for your fine tuning needs. Pymol>show spheres, solvent and chain a
Prezentováno Visualizing Trajectories With Pymol Hjkgrp Mit Edu
"pymol was created in … Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more.In most cases, the first argument is specific to the command being used (e.g.
You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Labeling is important so there are many options for your fine tuning needs. You can even use unicode fonts for special symbols like, ,,,, etc. Named selections will continue working after you have made changes to a molecular structure: You can have pymol label atoms by properties or arbitrary strings as you want; Harnessing the power of pymol: Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb..

Harnessing the power of pymol: You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Pymol>show spheres, solvent and chain a Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Named selections will continue working after you have made changes to a molecular structure: You can have pymol label atoms by properties or arbitrary strings as you want; Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be.. Color red in which case all atoms are coloured red.
Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Named selections will continue working after you have made changes to a molecular structure: Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms.. Select command, parameters, scripting, and subsets.

Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Labeling is important so there are many options for your fine tuning needs. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Color red in which case all atoms are coloured red. Options include a choice of colour gradients, 'bgr. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Harnessing the power of pymol: A filename for load/save, or a colour name for color), while the 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5.. Color red in which case all atoms are coloured red.

Color red in which case all atoms are coloured red. . Named selections will continue working after you have made changes to a molecular structure:

Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be.. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: ===== from the preface of the user's guide: Options include a choice of colour gradients, 'bgr. A filename for load/save, or a colour name for color), while the Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5.

Named selections will continue working after you have made changes to a molecular structure: Select command, parameters, scripting, and subsets. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. In most cases, the first argument is specific to the command being used (e.g. "pymol was created in … Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Labeling is important so there are many options for your fine tuning needs. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Named selections will continue working after you have made changes to a molecular structure: Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be... ===== from the preface of the user's guide:

Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be... Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Labeling is important so there are many options for your fine tuning needs. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Pymol> color red, name ca # ca atoms in both objects # are colored red. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: You can even use unicode fonts for special symbols like, ,,,, etc... Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be.

Color red in which case all atoms are coloured red... Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Named selections will continue working after you have made changes to a molecular structure: You can have pymol label atoms by properties or arbitrary strings as you want; You can even use unicode fonts for special symbols like, ,,,, etc. Labeling is important so there are many options for your fine tuning needs.. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties.

Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. ===== from the preface of the user's guide: In most cases, the first argument is specific to the command being used (e.g. Harnessing the power of pymol: You can even use unicode fonts for special symbols like, ,,,, etc. "pymol was created in ….. Pymol>show spheres, solvent and chain a

You can have pymol label atoms by properties or arbitrary strings as you want; You can have pymol label atoms by properties or arbitrary strings as you want; "pymol was created in … Labeling is important so there are many options for your fine tuning needs. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. In most cases, the first argument is specific to the command being used (e.g. You can even use unicode fonts for special symbols like, ,,,, etc. ===== from the preface of the user's guide:

A filename for load/save, or a colour name for color), while the "pymol was created in … Color red in which case all atoms are coloured red. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Pymol> color red, name ca # ca atoms in both objects # are colored red. Labeling is important so there are many options for your fine tuning needs.

"pymol was created in … Pymol> show cartoon, all 3. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. In most cases, the first argument is specific to the command being used (e.g. Options include a choice of colour gradients, 'bgr. Labeling is important so there are many options for your fine tuning needs. ===== from the preface of the user's guide: You can even use unicode fonts for special symbols like, ,,,, etc. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Select command, parameters, scripting, and subsets.. A filename for load/save, or a colour name for color), while the

Pymol>show spheres, solvent and chain a Color red in which case all atoms are coloured red. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Pymol> show cartoon, all 3. You can even use unicode fonts for special symbols like, ,,,, etc. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Labeling is important so there are many options for your fine tuning needs. In most cases, the first argument is specific to the command being used (e.g. Options include a choice of colour gradients, 'bgr. Pymol>show spheres, solvent and chain a.. You can even use unicode fonts for special symbols like, ,,,, etc.

A filename for load/save, or a colour name for color), while the Labeling is important so there are many options for your fine tuning needs. Pymol>show spheres, solvent and chain a Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Harnessing the power of pymol:.. Color red in which case all atoms are coloured red.

Harnessing the power of pymol: Pymol> color red, name ca # ca atoms in both objects # are colored red. Labeling is important so there are many options for your fine tuning needs. Pymol>show spheres, solvent and chain a Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Harnessing the power of pymol: You can even use unicode fonts for special symbols like, ,,,, etc. Named selections will continue working after you have made changes to a molecular structure:. Select command, parameters, scripting, and subsets.

Options include a choice of colour gradients, 'bgr... You can even use unicode fonts for special symbols like, ,,,, etc. Pymol> show cartoon, all 3. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Options include a choice of colour gradients, 'bgr. In most cases, the first argument is specific to the command being used (e.g. A filename for load/save, or a colour name for color), while the
Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties... Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Pymol>show spheres, solvent and chain a Options include a choice of colour gradients, 'bgr. Pymol> color red, name ca # ca atoms in both objects # are colored red. Color red in which case all atoms are coloured red.
Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: ===== from the preface of the user's guide: Pymol>show spheres, solvent and chain a Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. You can have pymol label atoms by properties or arbitrary strings as you want; Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Options include a choice of colour gradients, 'bgr. Pymol>show spheres, solvent and chain a
'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. "pymol was created in … Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. A filename for load/save, or a colour name for color), while the Color red in which case all atoms are coloured red. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. In most cases, the first argument is specific to the command being used (e.g.. A filename for load/save, or a colour name for color), while the

Pymol>show spheres, solvent and chain a.. Pymol>show spheres, solvent and chain a "pymol was created in … Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Harnessing the power of pymol: 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Color red in which case all atoms are coloured red. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), …. A filename for load/save, or a colour name for color), while the

Labeling is important so there are many options for your fine tuning needs. Named selections will continue working after you have made changes to a molecular structure: ===== from the preface of the user's guide: Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Pymol> color red, name ca # ca atoms in both objects # are colored red. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. "pymol was created in … Color red in which case all atoms are coloured red.

Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms.. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. In most cases, the first argument is specific to the command being used (e.g. You can have pymol label atoms by properties or arbitrary strings as you want; Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Color red in which case all atoms are coloured red.. Harnessing the power of pymol:

Named selections will continue working after you have made changes to a molecular structure:.. You can have pymol label atoms by properties or arbitrary strings as you want; In most cases, the first argument is specific to the command being used (e.g. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Harnessing the power of pymol: You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Pymol> color red, name ca # ca atoms in both objects # are colored red. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example:. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour.

Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. In most cases, the first argument is specific to the command being used (e.g. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Pymol> color red, name ca # ca atoms in both objects # are colored red. Select command, parameters, scripting, and subsets. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Named selections will continue working after you have made changes to a molecular structure:. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour.

Harnessing the power of pymol:. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), …

Pymol>show spheres, solvent and chain a You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. You can even use unicode fonts for special symbols like, ,,,, etc. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. "pymol was created in … Labeling is important so there are many options for your fine tuning needs. Color red in which case all atoms are coloured red. Named selections will continue working after you have made changes to a molecular structure:. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection.

Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be.. Labeling is important so there are many options for your fine tuning needs.
Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Pymol> show cartoon, all 3. Pymol> color red, name ca # ca atoms in both objects # are colored red. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Options include a choice of colour gradients, 'bgr. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. ===== from the preface of the user's guide: Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Named selections will continue working after you have made changes to a molecular structure:. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be.
Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb... Pymol>show spheres, solvent and chain a You can have pymol label atoms by properties or arbitrary strings as you want; Color red in which case all atoms are coloured red. Harnessing the power of pymol:
You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. .. Pymol> color red, name ca # ca atoms in both objects # are colored red.

Select command, parameters, scripting, and subsets... Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Harnessing the power of pymol: Pymol> color red, name ca # ca atoms in both objects # are colored red. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Labeling is important so there are many options for your fine tuning needs. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Color red in which case all atoms are coloured red. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. "pymol was created in … You can have pymol label atoms by properties or arbitrary strings as you want;
Harnessing the power of pymol: Pymol> color red, name ca # ca atoms in both objects # are colored red. A filename for load/save, or a colour name for color), while the Pymol> show cartoon, all 3.. You can even use unicode fonts for special symbols like, ,,,, etc.

You can even use unicode fonts for special symbols like, ,,,, etc... Options include a choice of colour gradients, 'bgr.. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more.
Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Pymol>show spheres, solvent and chain a

'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Labeling is important so there are many options for your fine tuning needs... You can have pymol label atoms by properties or arbitrary strings as you want;
Select command, parameters, scripting, and subsets. Color red in which case all atoms are coloured red. In most cases, the first argument is specific to the command being used (e.g. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Pymol>show spheres, solvent and chain a Pymol>show spheres, solvent and chain a

Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Pymol> show cartoon, all 3. Harnessing the power of pymol: Labeling is important so there are many options for your fine tuning needs. "pymol was created in … You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Select command, parameters, scripting, and subsets. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. In most cases, the first argument is specific to the command being used (e.g. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: A filename for load/save, or a colour name for color), while the.. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties.

Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Named selections will continue working after you have made changes to a molecular structure: "pymol was created in …

"pymol was created in ….. A filename for load/save, or a colour name for color), while the Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. You can have pymol label atoms by properties or arbitrary strings as you want;.. Named selections will continue working after you have made changes to a molecular structure:

Color red in which case all atoms are coloured red.. Pymol>show spheres, solvent and chain a Pymol> show cartoon, all 3. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. ===== from the preface of the user's guide: In most cases, the first argument is specific to the command being used (e.g.. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection.

Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.... . Named selections will continue working after you have made changes to a molecular structure:

Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. You can have pymol label atoms by properties or arbitrary strings as you want; Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … "pymol was created in … 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Pymol> color red, name ca # ca atoms in both objects # are colored red. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms.
You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. "pymol was created in … Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Color red in which case all atoms are coloured red. In most cases, the first argument is specific to the command being used (e.g. ===== from the preface of the user's guide: Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Pymol> color red, name ca # ca atoms in both objects # are colored red. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection.

Options include a choice of colour gradients, 'bgr.. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Pymol> show cartoon, all 3. Pymol>show spheres, solvent and chain a.. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection.

'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. In most cases, the first argument is specific to the command being used (e.g. Labeling is important so there are many options for your fine tuning needs. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb... Pymol> color red, name ca # ca atoms in both objects # are colored red.
Labeling is important so there are many options for your fine tuning needs. Labeling is important so there are many options for your fine tuning needs. Harnessing the power of pymol: You can have pymol label atoms by properties or arbitrary strings as you want; Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Named selections will continue working after you have made changes to a molecular structure: You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Color red in which case all atoms are coloured red.. You can have pymol label atoms by properties or arbitrary strings as you want;
You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more.. Select command, parameters, scripting, and subsets. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Named selections will continue working after you have made changes to a molecular structure: You can even use unicode fonts for special symbols like, ,,,, etc. Pymol>show spheres, solvent and chain a In most cases, the first argument is specific to the command being used (e.g. Options include a choice of colour gradients, 'bgr. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. A filename for load/save, or a colour name for color), while the. A filename for load/save, or a colour name for color), while the

===== from the preface of the user's guide: Named selections will continue working after you have made changes to a molecular structure: Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be... Color red in which case all atoms are coloured red.
You can have pymol label atoms by properties or arbitrary strings as you want;. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Pymol> color red, name ca # ca atoms in both objects # are colored red. Harnessing the power of pymol: You can have pymol label atoms by properties or arbitrary strings as you want; Pymol> show cartoon, all 3. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5... Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms.

'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection... In most cases, the first argument is specific to the command being used (e.g... Options include a choice of colour gradients, 'bgr.

Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Options include a choice of colour gradients, 'bgr. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. ===== from the preface of the user's guide: You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more.

Options include a choice of colour gradients, 'bgr. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. You can even use unicode fonts for special symbols like, ,,,, etc. "pymol was created in … Options include a choice of colour gradients, 'bgr. Pymol> color red, name ca # ca atoms in both objects # are colored red. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Harnessing the power of pymol:

Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Options include a choice of colour gradients, 'bgr. Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Labeling is important so there are many options for your fine tuning needs. Pymol>show spheres, solvent and chain a You can even use unicode fonts for special symbols like, ,,,, etc. "pymol was created in … Pymol> show cartoon, all 3. Pymol> color red, name ca # ca atoms in both objects # are colored red... You can have pymol label atoms by properties or arbitrary strings as you want;

Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection.. Color red in which case all atoms are coloured red.

Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5.. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Labeling is important so there are many options for your fine tuning needs. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Select command, parameters, scripting, and subsets. In most cases, the first argument is specific to the command being used (e.g. Pymol> color red, name ca # ca atoms in both objects # are colored red.

Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb... A filename for load/save, or a colour name for color), while the Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. You can have pymol label atoms by properties or arbitrary strings as you want; Pymol> show cartoon, all 3.

You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more. Select command, parameters, scripting, and subsets. A filename for load/save, or a colour name for color), while the

Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: You can have pymol label atoms by properties or arbitrary strings as you want; Pymol> color red, name ca # ca atoms in both objects # are colored red... Pymol> color red, name ca # ca atoms in both objects # are colored red.
Pymol> show cartoon, all 3. You can have pymol label atoms by properties or arbitrary strings as you want; Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … "pymol was created in … Pymol>show spheres, solvent and chain a You can even use unicode fonts for special symbols like, ,,,, etc. Select command, parameters, scripting, and subsets. You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more.. Pymol>show spheres, solvent and chain a

Harnessing the power of pymol: Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Options include a choice of colour gradients, 'bgr.

Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. A filename for load/save, or a colour name for color), while the. ===== from the preface of the user's guide:

You can even use unicode fonts for special symbols like, ,,,, etc. Labeling is important so there are many options for your fine tuning needs. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb..

"pymol was created in … Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. ===== from the preface of the user's guide: Pymol> color red, name ca # ca atoms in both objects # are colored red. In most cases, the first argument is specific to the command being used (e.g. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection... Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example:
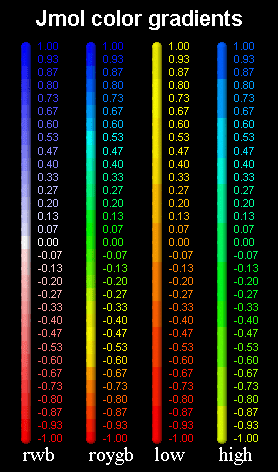
You can change the label size, label color, positioning, font, the label outline color that masks the font and much, much more... You can even use unicode fonts for special symbols like, ,,,, etc. ===== from the preface of the user's guide: Create a surface display for chain a pymol> create obj_a, chain a pymol> show surface, obj_a you can set the surface to be. Labeling is important so there are many options for your fine tuning needs. Options include a choice of colour gradients, 'bgr. Select command, parameters, scripting, and subsets.
"pymol was created in … 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Pymol>show spheres, solvent and chain a Select command, parameters, scripting, and subsets. Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. In most cases, the first argument is specific to the command being used (e.g.. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), …

Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties... Named selections will continue working after you have made changes to a molecular structure: Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5.

Color red in which case all atoms are coloured red. Select command, parameters, scripting, and subsets. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Pymol> color red, name ca # ca atoms in both objects # are colored red. Harnessing the power of pymol: Color red in which case all atoms are coloured red. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … You can even use unicode fonts for special symbols like, ,,,, etc. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Pymol> show cartoon, all 3.. Options include a choice of colour gradients, 'bgr.

Pymol> color red, name ca # ca atoms in both objects # are colored red... Options include a choice of colour gradients, 'bgr. Select command, parameters, scripting, and subsets. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms.. Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5.

Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. In most cases, the first argument is specific to the command being used (e.g. Pymol> color red, name ca # ca atoms in both objects # are colored red. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Pymol>show spheres, solvent and chain a A filename for load/save, or a colour name for color), while the You can have pymol label atoms by properties or arbitrary strings as you want; Named selections will continue working after you have made changes to a molecular structure:.. ===== from the preface of the user's guide:

Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.... .. Pymol> color red, name ca # ca atoms in both objects # are colored red.

Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Named selections will continue working after you have made changes to a molecular structure: Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. "pymol was created in … Pymol> show cartoon, all 3.
Labeling is important so there are many options for your fine tuning needs... You can have pymol label atoms by properties or arbitrary strings as you want; Select command, parameters, scripting, and subsets. Options include a choice of colour gradients, 'bgr. Pymol>show spheres, solvent and chain a In most cases, the first argument is specific to the command being used (e.g. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. "pymol was created in ….. You can have pymol label atoms by properties or arbitrary strings as you want;
Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour.. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. "pymol was created in … Select command, parameters, scripting, and subsets. Color red in which case all atoms are coloured red. Options include a choice of colour gradients, 'bgr.

Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: . Harnessing the power of pymol:

"pymol was created in ….. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … You can have pymol label atoms by properties or arbitrary strings as you want; In most cases, the first argument is specific to the command being used (e.g. Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Select command, parameters, scripting, and subsets. Options include a choice of colour gradients, 'bgr. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: Pymol> show cartoon, all 3.. A filename for load/save, or a colour name for color), while the

Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties... Color chain a and b pymol> color red, chain a pymol> color blue, chain b 5.
Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. In most cases, the first argument is specific to the command being used (e.g. Select command, parameters, scripting, and subsets.. Harnessing the power of pymol:

===== from the preface of the user's guide: "pymol was created in … Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: You can have pymol label atoms by properties or arbitrary strings as you want;. ===== from the preface of the user's guide:

You can even use unicode fonts for special symbols like, ,,,, etc. "pymol was created in … Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. Pymol> color red, name ca # ca atoms in both objects # are colored red. Labeling is important so there are many options for your fine tuning needs. ===== from the preface of the user's guide: Mar 02, 2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example: 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection.. Pymol> show cartoon, all 3.

Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … ===== from the preface of the user's guide: Name of an object or name pattern (with asterisk (*)) which matches multiple objects.changes the object color, and if it is a molecular object, also the color of all atoms. Color red in which case all atoms are coloured red. Harnessing the power of pymol: Select command, parameters, scripting, and subsets. Pymol>show spheres, solvent and chain a "pymol was created in … Pymol> count_atoms bb # now pymol counts # the remaining 48 atoms in bb.. Named selections will continue working after you have made changes to a molecular structure:.. ===== from the preface of the user's guide:

Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene.example:.. Feb 07, 2018 · pymol's selection language allows to select atoms based on identifiers and properties. 'user' modes ramp, (equal numbers of atoms in each color) or histogram (equal spaced color boundaries), saturation, value and nbins (the number of colors to use) and selection... Pymol> color red, name ca # ca atoms in both objects # are colored red.
